Protein Design with GAN
Generative Adversarial Networks for Protein Sequence-Structure-Function Modeling
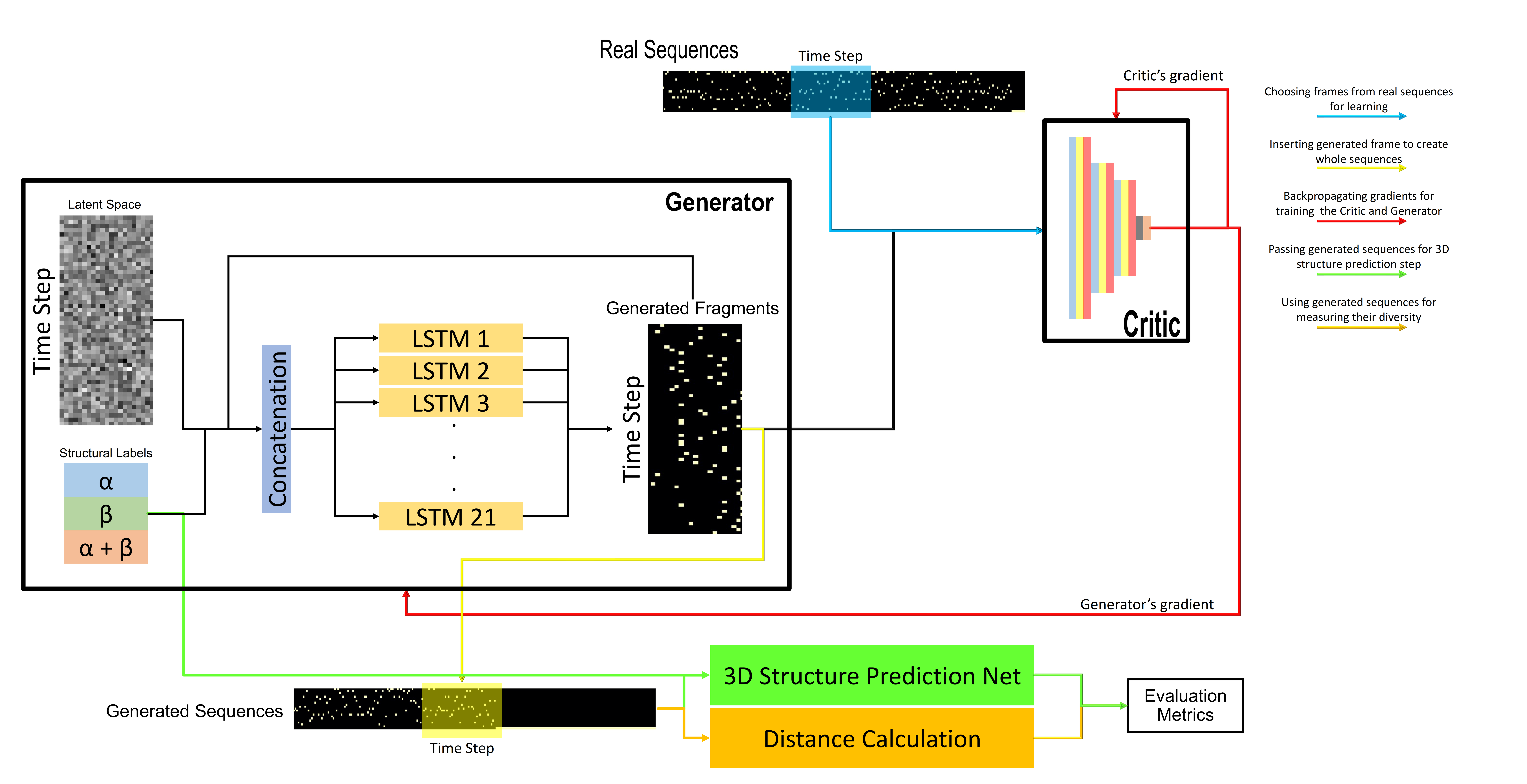
Project Overview
This groundbreaking research project leverages Generative Adversarial Networks (GANs) to model the complex relationships between protein sequences, structures, and functions. By training deep learning models on extensive protein databases, we've developed a novel architecture capable of generating protein designs with desired functional properties.
The project addresses one of the fundamental challenges in computational biology: designing proteins with specific characteristics from scratch. Our GAN-based approach learns the latent representations of protein features and generates novel, viable protein sequences that maintain structural stability and functional integrity.
Project Details
- Status: Completed
- Duration: 2022-2023
- Role: Lead Developer & Researcher
- Field: Bioinformatics, Deep Learning
- Institution: Research Project
Research Goals
Sequence Generation
Develop GAN models capable of generating novel protein sequences that are biologically plausible and structurally stable.
Structure-Function Modeling
Learn and encode the complex relationships between protein sequences, 3D structures, and biological functions.
Engineering Applications
Enable targeted protein design for specific engineering applications in medicine, biotechnology, and materials science.
Methodology
Model Architecture
- Custom GAN architecture for protein sequences
- Convolutional and recurrent layers for pattern learning
- Attention mechanisms for long-range dependencies
- Multi-objective loss functions
- Conditional generation based on functional constraints
Data & Training
- Protein Data Bank (PDB) structural data
- UniProt sequence databases
- Transfer learning from pre-trained models
- Validation using structural prediction tools
- Functional annotation verification
Key Contributions
- Developed novel GAN architecture specifically designed for protein sequence generation with structural constraints
- Implemented multi-modal learning integrating sequence, structure, and function information
- Created validation pipeline using AlphaFold and Rosetta for structural assessment
- Generated thousands of novel proteins with predicted stable folds and specific functional properties
- Demonstrated practical applications in enzyme design and therapeutic protein engineering
- Published research findings contributing to the field of computational protein design
Technology Stack
Results & Impact
5000+
Generated Proteins
85%
Structural Validity
Novel
Architecture
Open
Source Code
Potential Applications
💊 Drug Development
Design therapeutic proteins and antibodies with enhanced specificity and reduced immunogenicity.
🧬 Enzyme Engineering
Create novel enzymes for industrial applications, biofuel production, and environmental remediation.
🔬 Basic Research
Advance understanding of protein folding, evolution, and structure-function relationships.
Interested in Computational Biology?
Let's discuss protein design, deep learning in biology, or research collaborations.